
- #Chromatogram viewer for mac install#
- #Chromatogram viewer for mac software#
- #Chromatogram viewer for mac download#
If you receive files from our 3100 and wish to open them on a Macintosh computer, you must first convert them so they are readable on the Mac.
#Chromatogram viewer for mac software#
In addition, there are numerous commercial software packages that will allow you to open the ABI files and usually provide many more options such as alignment, contig building, potential open reading frames, etc.
#Chromatogram viewer for mac download#
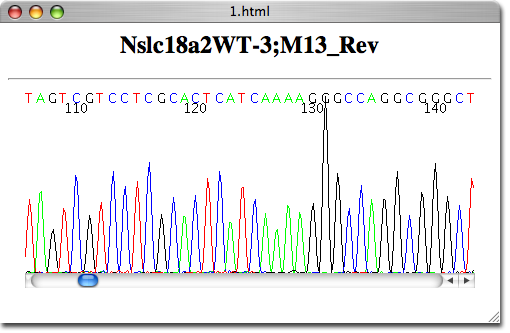
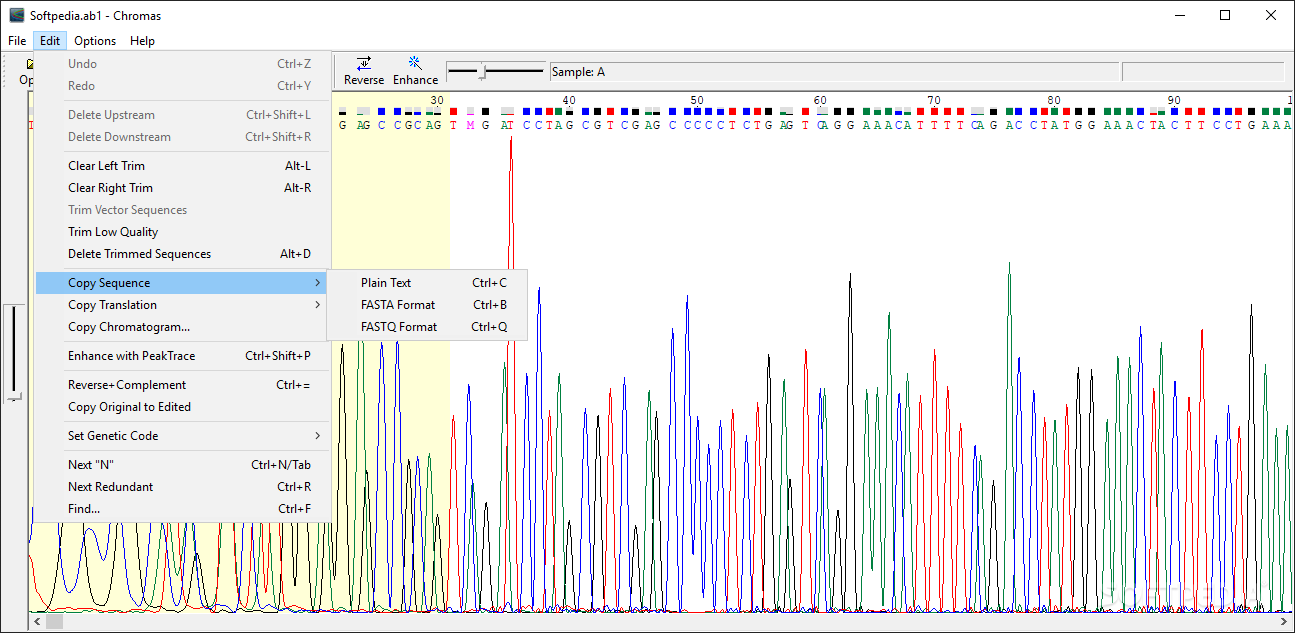
There are a few freeware programs, for both Mac and PC, which you can download from the Internet that will allow you to view, edit and print your color trace files.
#Chromatogram viewer for mac install#
A wizard is available for each workflow sample.In order to open and view the ABI chromatogram files we now provide for downloading from our server, you must install a program on your computer that will allow you to do that. There is an embedded library of workflow samples to convert, filter, and annotate data, with several pipelines to analyze NGS data developed in collaboration with NIH NIAID. The graphical interface also allows controlling the workflow execution, storing the parameters, and so on. This allows their reuse, and transfer between users.Ī workflow can be run using the graphical interface or launched from the command line. Workflows are stored in a special text format. The elements can be based on a command-line tool or a script. Using Workflow Designer also allows creating custom workflow elements. The elements that a workflow consists of correspond to the bulk of algorithms integrated into UGENE. It helps to avoid data transfer issues, whereas other tools’ reliance on remote file storage and internet connectivity does not. The distinguishing feature of Workflow Designer, relative to other bioinformatics workflow management systems is that workflows are executed on a local computer. UGENE Workflow Designer allows creating and running complex computational workflow schemas.
Multiple sequence alignment: Clustal W and O, MUSCLE, Kalign, MAFFT, T-Coffee.Create, edit, and annotate nucleic acid and protein sequences.To improve performance, UGENE uses multi-core processors (CPUs) and graphics processing units (GPUs) to optimize a few algorithms. A set of sample workflows is available in the Workflow Designer, to annotate sequences, convert data formats, analyze NGS data, etc.īeside the graphical interface, UGENE also has a command-line interface. Blocks can be created with command line tools or a script. The workflow consists of blocks such as data readers, blocks executing embedded tools and algorithms, and data writers. Using UGENE Workflow Designer, it is possible to streamline a multi-step analysis. UGENE provides a graphical user interface (GUI) for the pre-built tools so biologists with no computer programming skills can access those tools more easily. UGENE integrates dozens of well-known biological tools, algorithms, and original tools in the context of genomics, evolutionary biology, virology, and other branches of life science. The data can be stored both locally (on a personal computer) and on a shared storage (e.g., a lab database). UGENE helps biologists to analyze various biological genetics data, such as sequences, annotations, multiple alignments, phylogenetic trees, NGS assemblies, and others.


 0 kommentar(er)
0 kommentar(er)
